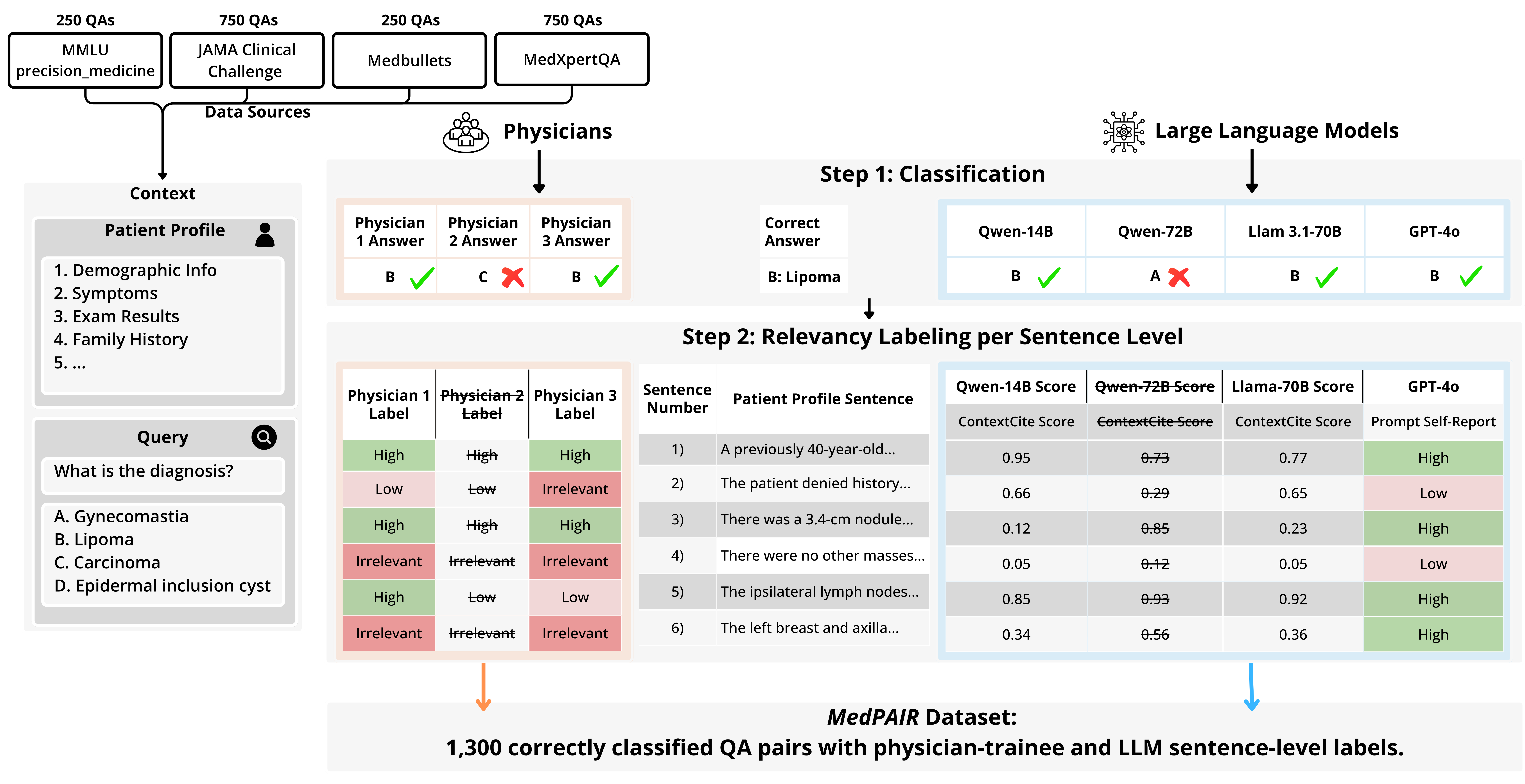

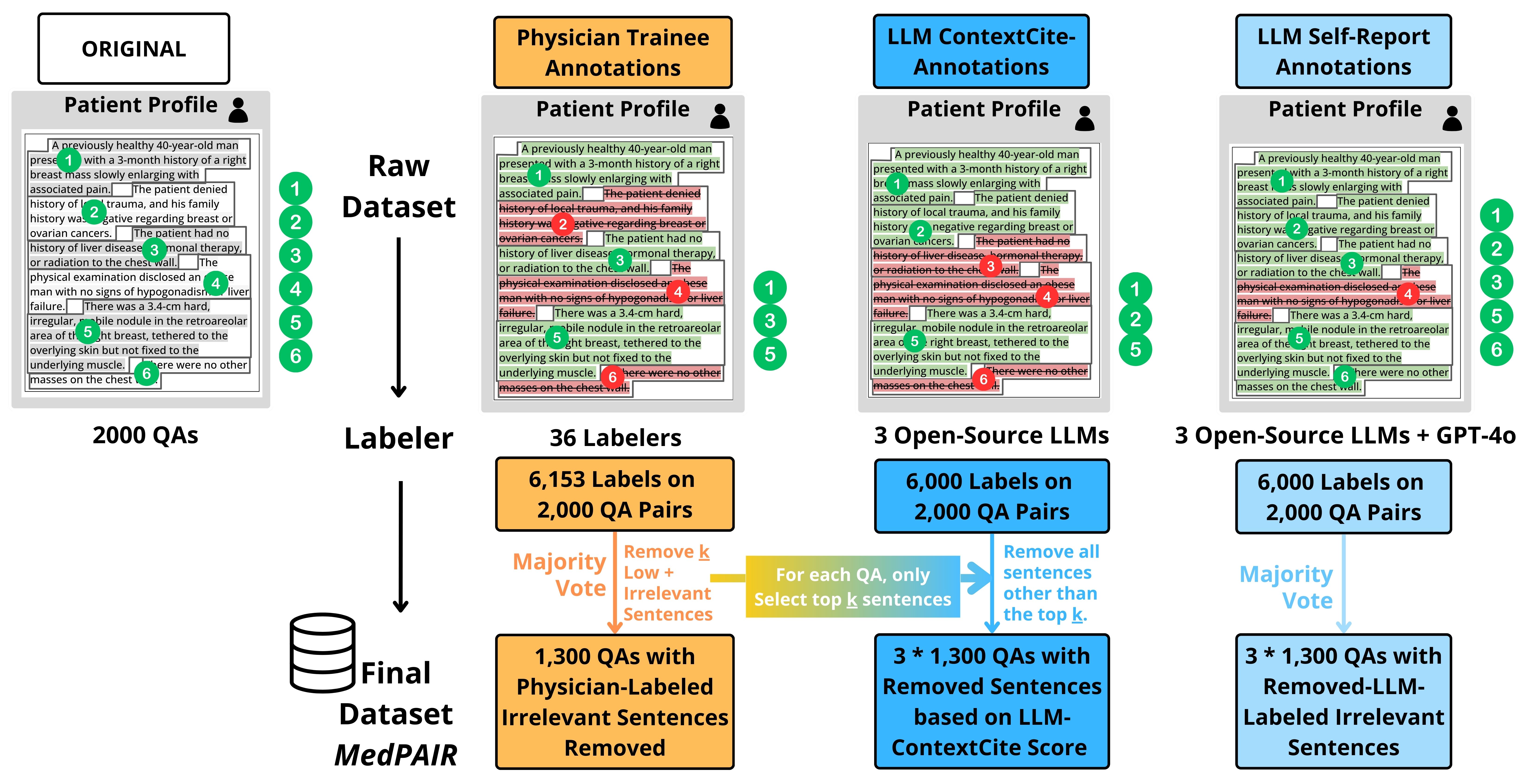
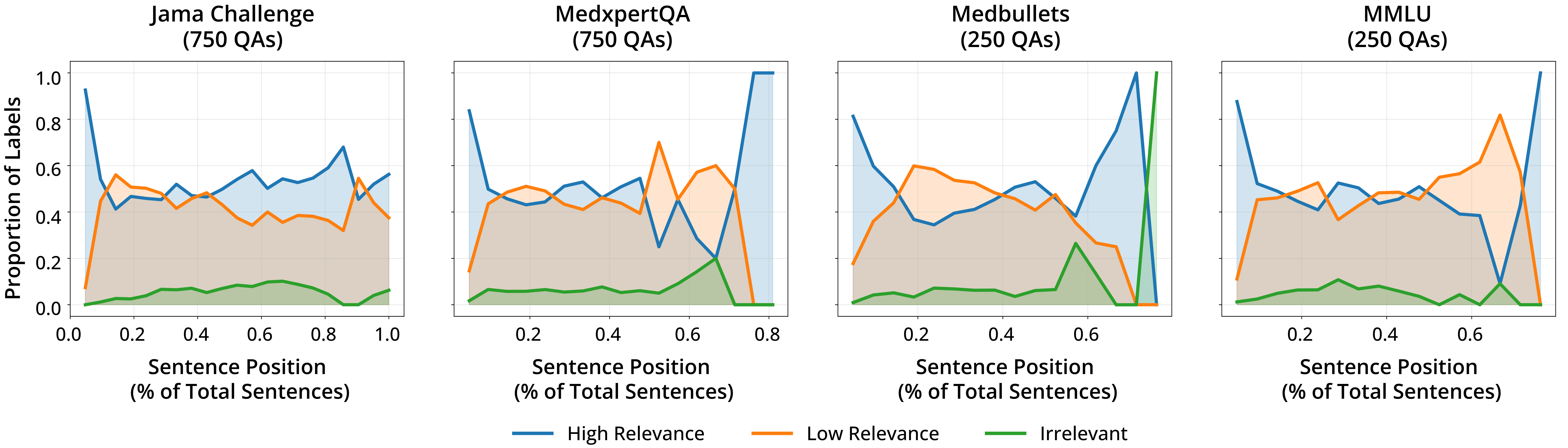
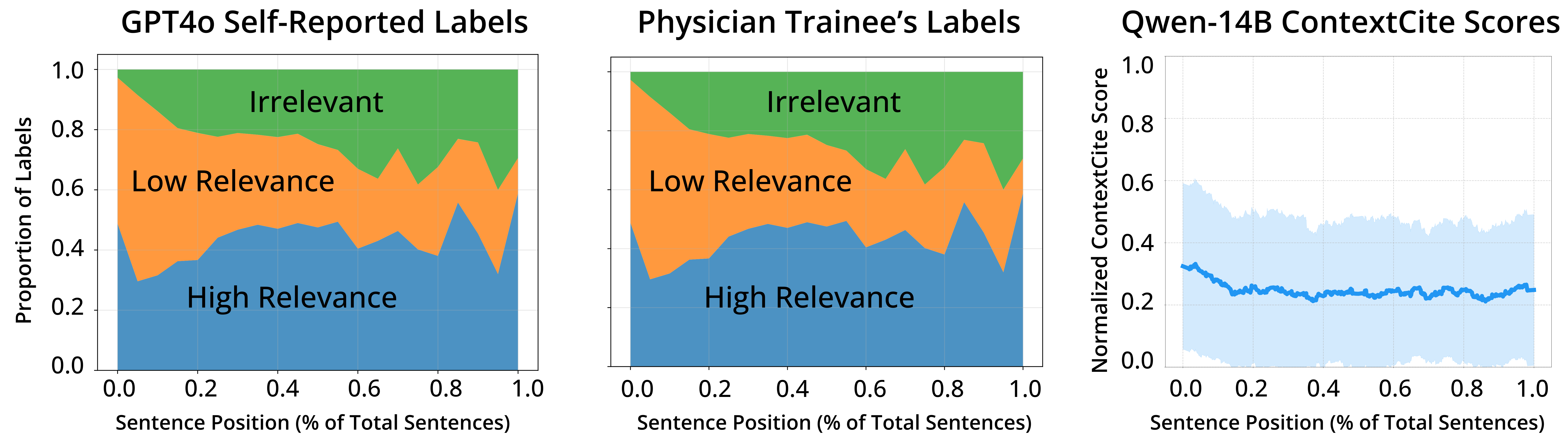
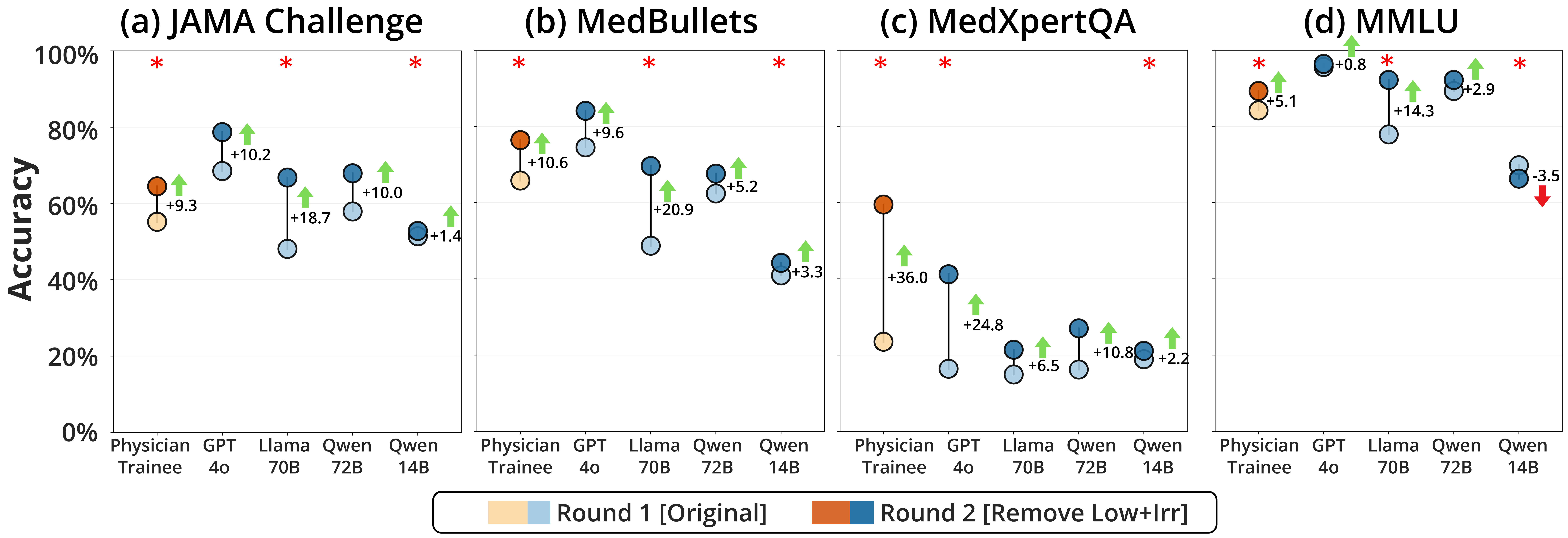
Large Language Models (LLMs) have demonstrated remarkable performance on various medical question-answering (QA) benchmarks, including standardized medical exams. However, correct answers alone do not ensure correct logic, and models may reach accurate conclusions through flawed processes. In this study, we introduce the comparative reasoning benchmark MedPAIR (Medical Physicians and AI Relevance Alignment in Medical Question Answering) to evaluate how physicians and LLMs prioritize relevant information when answering QA questions. We obtain annotations on 2,000 QA pairs from 36 physicians, labeling each sentence within the question components for relevancy. We compare these relevancy estimates to those for LLMs, and further evaluate the impact of these ``relevant'' subsets on downstream task performance for both humans and LLMs. We further implemented a structured intervention where physicians guided LLMs to focus on relevant information. We find that LLMs are frequently not aligned with physicians' estimates of content relevancy. Further, we highlight the limitations of current LLM behavior and the importance of aligning AI reasoning with clinical standards.
@misc{hao2025medpairmeasuringphysiciansai,
title={MedPAIR: Measuring Physicians and AI Relevance Alignment in Medical Question Answering},
author={Yuexing Hao and Kumail Alhamoud and Hyewon Jeong and Haoran Zhang and Isha Puri and Philip Torr and Mike Schaekermann and Ariel D. Stern and Marzyeh Ghassemi},
year={2025},
eprint={2505.24040},
archivePrefix={arXiv},
primaryClass={cs.CL},
url={https://arxiv.org/abs/2505.24040},
}